The genetic codes
 Since the discovery that DNA is a duplex molecule [1] which stores and replicates the information of living systems, the goal of modern biology has been to understand how the blueprint of a living system is encoded in its DNA. The first quest was to learn how DNA's four letter nucleotide code was translated into the 20 letter amino acid code of proteins. Experiments conducted in the 1960’s revealed that different combinations of triplet DNA bases encoded specific amino acids to produce the “universal” genetic code, which is nearly identical in all species that have been examined to date [2].
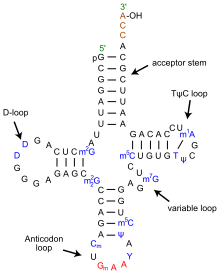
Since the discovery that DNA is a duplex molecule [1] which stores and replicates the information of living systems, the goal of modern biology has been to understand how the blueprint of a living system is encoded in its DNA. The first quest was to learn how DNA's four letter nucleotide code was translated into the 20 letter amino acid code of proteins. Experiments conducted in the 1960’s revealed that different combinations of triplet DNA bases encoded specific amino acids to produce the “universal” genetic code, which is nearly identical in all species that have been examined to date [2]. Translating mRNA into protein is a complex process, however, that involves many proteins and ribosomal RNA (rRNA) collectively organized in ribosomes. As the ribosomes read the mRNA sequence, transfer RNA (tRNA) molecules bring individual amino acids to the ribosome where they are added to a growing polypeptide chain. The universal genetic code explained how tri-nucleotide sequences specified amino acids. It could also be used to elucidate the anti-codon portion of tRNA [3], but it could not explain how the correct amino acid was added to the tRNA. For that another genetic code needed to be cracked. In this code, first proposed in 1988 [4], multiple sequences, including the anti-codon loop, within each tRNA molecule are recognized by a matched enzyme that combines an amino acid with its appropriate tRNA.
Codes to create diversity
The above codes are involved with the process of translating genetic sequences into protein. Most eukaryotic genes, and a few prokaryotic genes, cannot be translated in a continuous way because the protein coding regions (exons) are interrupted by non-coding regions (introns). When DNA is first transcribed into RNA, all regions are included and the introns must be excised to form the final messenger RNA (mRNA). This process makes it possible to create many different proteins from a single gene through alternative splicing in which exons are either differentially removed or portions of exons are joined together. Alternative splicing occurs in development and tissue specific ways; many disease causing mutations disrupt splicing patterns. So, understanding the codes that control splicing is an important research topic.
Some of the splicing codes, such as the exon boundaries, are well known, and others are not. In “Deciphering the Splicing Code,” Barash and colleagues looked at thousands of alternatively spliced exons - and surrounding intron sequences - from 27 mouse tissues to unravel over 1000 sequence features that could define a new genetic code. Their goal is build catalogs of motifs that could be used to predict splicing patterns of uncharacterized exons and determine how mutations might affect splicing.
Codes to create diversity
The above codes are involved with the process of translating genetic sequences into protein. Most eukaryotic genes, and a few prokaryotic genes, cannot be translated in a continuous way because the protein coding regions (exons) are interrupted by non-coding regions (introns). When DNA is first transcribed into RNA, all regions are included and the introns must be excised to form the final messenger RNA (mRNA). This process makes it possible to create many different proteins from a single gene through alternative splicing in which exons are either differentially removed or portions of exons are joined together. Alternative splicing occurs in development and tissue specific ways; many disease causing mutations disrupt splicing patterns. So, understanding the codes that control splicing is an important research topic.
Some of the splicing codes, such as the exon boundaries, are well known, and others are not. In “Deciphering the Splicing Code,” Barash and colleagues looked at thousands of alternatively spliced exons - and surrounding intron sequences - from 27 mouse tissues to unravel over 1000 sequence features that could define a new genetic code. Their goal is build catalogs of motifs that could be used to predict splicing patterns of uncharacterized exons and determine how mutations might affect splicing.
Using data from existing microarray experiments, RNA sequence features compiled from the literature, and other known attributes of RNA structure, Barash and co-workers developed computer models to determine which combinations of features best correlated with experimental observations. The resulting computer program provided tissue specific splicing predictions of whether an exon would be included or excluded based on its surrounding motif sequences and tissue type with reasonable success. More importantly, the program could be used to identify interaction networks that identified pairs of motifs that were frequently observed together.
Predicting alternative splicing is at an early stage, but as pointed out be the editorial summary, the approach of Barash and co-workers will be improved by the massive amounts of data being generated by new sequencing technologies and applications like RNA-Seq and various protein binding assays. The real test will be expanding the models to new tissues and human genomics. In the meantime, if you want to test their models on some of your data or explore new regulatory elements, the Frey lab has developed a web tool that can be accessed at http://genes.toronto.edu/wasp/.
I’m done with seconds, can I have a third?
As an aside, the authors of the editorial summary coined the work as the second genetic code. I find this amusing, because this would be the third second genetic code. The aminoacyl tRNA code was also coined the second genetic code, but people must have forgotten that, because another second genetic code was proposed in 2001. This genetic code describes how methylated DNA sequences regulate chromatin structure and gene regulation. Rather than have a third second genetic code, maybe we should refer to this as the third genetic code or the next generation code.
Further Reading
1. Watson JD, and Crick F (1953). "A structure for deoxyribose nucleic acid". Nature 171: 737–8.
I’m done with seconds, can I have a third?
As an aside, the authors of the editorial summary coined the work as the second genetic code. I find this amusing, because this would be the third second genetic code. The aminoacyl tRNA code was also coined the second genetic code, but people must have forgotten that, because another second genetic code was proposed in 2001. This genetic code describes how methylated DNA sequences regulate chromatin structure and gene regulation. Rather than have a third second genetic code, maybe we should refer to this as the third genetic code or the next generation code.
Further Reading
1. Watson JD, and Crick F (1953). "A structure for deoxyribose nucleic acid". Nature 171: 737–8.
2. http://en.wikipedia.org/wiki/Genetic_code
3. http://nobelprize.org/nobel_prizes/medicine/laureates/1968/holley-lecture.pdf
4. Hou YM, Schimmel P (1988) "A simple structural feature is a major determinant of the identity of a transfer RNA." Nature 333:140-5.

No comments:
Post a Comment