Next week, Nov 12-18, Super Computing comes to Seattle. On Wed, Nov 15 at 12:15-1:15 pm, @finchtalk (me) will host a Birds-of-a-Feater session on "Technologies for Managing BioData" in room TCC305.
I'll kick off the session by sharing stories from Geospiza's work experiences and the work of others. If you have a story to share please bring it. The session will provide an open platform. We plan to cover relational databases, HDF5 technologies, and NoSQL. If you want to join in because you are interested in learning, the abstract below will give you an idea of what will be discussed.
Abstract:
DNA sequencing and related technologies are producing tremendous volumes of data. The raw data from these instruments needs to be reduced through alignment or assembly into forms that can be further processed to yield scientifically or clinically actionable information. The entire data workflow process requires multiple programs and information resources. Standard formats and software tools that meet high performance computing requirements are lacking, but technical approaches are emerging. In this BoF, options such as BAM, BioHDF, VCF and other formats, and corresponding tools, will be reviewed for their utility in meeting a broad set of requirements. The goal of the BoF is look beyond DNA sequencing and discuss the requirements for data management technologies that can integrate sequence data with data collected from other platforms such as quantitative PCR, mass spectrometry, and imaging systems. We will also explore the technical requirements for working with data from large numbers of samples.
Finches continually evolve and so does science. Follow the conversation to learn how data impacts our world view.
Tuesday, November 8, 2011
Thursday, October 13, 2011
Personalities of Personal Genomes
"People say they want their genetic information, but they don’t." "The speaker's views of data return are frankly repugnant." These were some of the [paraphrased] comments and tweets expressed during Cold Spring Harbor's fourth annual conference entitled "Personal Genomes" held Sep 30 - Oct 2, 2011. The focus of which was to explore the latest technologies and approaches for sequencing genomes, exomes, and transcriptomes in the context of how genome science is, and will be, impacting clinical care.
The future may be close than we think
In previous years, the concept of personal genome sequencing as a way to influence medical treatment was a vision. Last year, the reality of the vision was evident through a limited number of examples. This year, several new examples were presented along with the establishment of institutional programs for genomic-based medicine. The driver being the continuing decreases in data collection costs combined with corresponding access to increasing amounts of data. According to Richard Gibbs (Baylor College of Medicine) we will have close to 5000 genomes completely sequenced by the end of this year and by the end of 2012, 30,000 complete genome sequences are expected.
The growth of genome sequencing is now significant enough that leading institutions are also beginning to establish guidelines for genomics-based medicine. Hence, an ethics panel discussion was held during the conference. The conversation about how DNA sequence data may be used has been an integral discussion since the beginning of the Genome Project. Indeed James Watson shared his lament for having to fund ethics research and directly asked the panel if they have done any good. There was a general consensus, from the panel, and audience members who have had their genomes sequenced, that ethics funding has helped by establishing genetic counseling and eduction practices.
However, as pointed out by some audience members, this ethics panel, like many others, focused too heavily on the risks for individuals and society having their genomic data. In my view, the discussion would have been more interesting and balanced if the panel included the individuals who are working outside of institutions with new approaches for understanding health. Organizations like 23andMe, Patients LIke Me, or the Genetic Alliance bring a very different and valuable perspective to the conversation.
Ethics was a fraction of the conference. The remaining talks at were organized into six sessions that covered personal cancer genomics, medically actionable genomics, personal genomes, rare diseases, and clinical implementations of personal genomics. The key messages from these presentations and posters was that, while genomics-based medical approaches have demonstrated success, much more research needs to be done before such approaches are mainstream.
For example, in the case of cancer genomics, whole genome sequences from tumor and normal cells can give a picture of point mutations and structural rearrangements, but these data need to be accompanied by exome sequences to get the high read depth needed to accurately detect the low levels of rare mutations that may be disregulating cell growth or conferring resistance to treatment. Yet, the resulting profiles of variants are still inadequate to fully understand the functional consequences of the mutations. For this, transcriptome profiling is needed, and that is just the start.
Once the data are collected they need to be processed in different ways, filtered, and compared within and between samples. Information from many specialized databases will be used in conjunction with statistical analyses to develop insights that can be validated through additional assays and measurements. Finally, a lab seeking to do this work, and return results back to patients, will also need to be certified, minimally by CLIA standards. For many groups this is significant undertaking, and good partners with experience and strong capabilities like PerkinElmer will be needed.
Further Reading
Nature Coverage, Oct 6 issue:
Genomes on prescription
Other news and information:
Other news and information:
Friday, September 30, 2011
Wednesday, August 10, 2011
Stitching Protein-Protien Interactions via DNA Sequencing
Back in 2008, when groups were realizing the power of NGS technologies, I entitled a post "Next Gen Sequencing is not Sequencing DNA" to make the point that massively parallel ultra-high throughput DNA sequencing could be used to for quantitative assays that can measure transcriptome expression, protein-DNA interactions, methylation patterns, and more. Stitch-seq can now be added to a growing list of assays that include RNA-Seq, DNAse-Seq, ChIP-Seq, or HITS-CLIP, and others.
Stitch-seq explores the interactome, a term used to describe how the molecules of a cell interact in networks to carryout life's biochemical activities. Understanding how these networks are controlled through genetics and environmental stimuli is critical in discovering biomarkers that can be used to stratify disease and target highly specific therapies. However, the interactome is complex; studying it requires that interactions can be identified at high scale.
Many interactome studies focus on proteins. Traditional approaches involve specially constructed gene reporter systems. For example, in the two-hybrid approach, a portion of a protein encoding gene is combined with a gene fragment containing a DNA binding domain of a transcription factor (bait). In another construct a different protein encoding region is combined with the RNA polymerase binding domain fragment of the same transcription factor (prey).
When the DNA constructs are expressed, interactions can be measure by gene expression. If the protein attached to the bait interacts with the protein attached to the prey, transcription is initiated at the reporter gene. When reporter genes confer growth on selective media, interacting protein encoding segments can be identified by isolating the DNA from growing cells and sequencing the DNA constructs.
Therein lies the rub
 Until now, interactome studies combined high-throughtput assays systems with low-throughput characterization systems that PCR amplified the individual constructs and characterized the DNA by Sanger sequencing. Yu and colleagues overcame this problem by devising a new strategy that put potential interacting domains on common DNA fragments, via "stitch-PCR" to prepare libraries that can easily be sequenced by NGS methods. Using this method the team was able to increase overall assay throughput by 42% and measure 1000s of interactions.
Until now, interactome studies combined high-throughtput assays systems with low-throughput characterization systems that PCR amplified the individual constructs and characterized the DNA by Sanger sequencing. Yu and colleagues overcame this problem by devising a new strategy that put potential interacting domains on common DNA fragments, via "stitch-PCR" to prepare libraries that can easily be sequenced by NGS methods. Using this method the team was able to increase overall assay throughput by 42% and measure 1000s of interactions.While still low-throughput relative to the kinds of numbers were used to on NGS, increasing the throughput of protein interaction assays is an important step toward making systems biology experiments more scalable. It also adds another Seq to our growing collection of Assay-Seq methods.
Yu, H., Tardivo, L., Tam, S., Weiner, E., Gebreab, F., Fan, C., Svrzikapa, N., Hirozane-Kishikawa, T., Rietman, E., Yang, X., Sahalie, J., Salehi-Ashtiani, K., Hao, T., Cusick, M., Hill, D., Roth, F., Braun, P., & Vidal, M. (2011). Next-generation sequencing to generate interactome datasets Nature Methods, 8 (6), 478-480 DOI: 10.1038/nmeth.1597
Friday, July 8, 2011
It's not just science and technology
PerkinElmer is committed to improving human and environmental health. Sometimes that extends beyond delivering commercial products and services to how we participate in our community.
Geospiza was proud to help the local community last Tuesday (7/5) by volunteering, through PerkinElmer's Corporate Social Responsibility program, to clean up the area around Lake Union after the Seattle's Fourth of July Celebration.
It's also great way to post a couple of pictures of some of the Geospiza team.
About the activity
After thousands celebrate the Fourth of July and the watch the fireworks, Seattle’s Lake Union can be more trampled fields of chip bags and shining mounds of beer cans than amber waves of grain and purple mountains majesty.
As a member of the biotech community based around Lake Union, Geospiza donned gardening gloves and joined our neighbors for the 5th of July Lake Union Cleanup, a post-fireworks cleaning frenzy hosted by Starbucks, Puget Sound Keeper Alliance and Seattle Public Utilities.
Last year’s inaugural event had more than 700 volunteers collecting 1.15 tons of garbage – one-third of which was recycled. The cleaning event grew out of the community-based efforts to save the fireworks. When the 2010 show was cancelled due to lack of corporate sponsorship, the community launched a fund drive which raised enough money in less than 24hrs.
From April through the end of July, every site across PerkinElmer, including Geospiza, participates in one “For The Better Day” event to fulfill our company mission to improve human and environmental health while providing employees with an opportunity to build stronger teams and communities where we work, live and play.
Geospiza was proud to help the local community last Tuesday (7/5) by volunteering, through PerkinElmer's Corporate Social Responsibility program, to clean up the area around Lake Union after the Seattle's Fourth of July Celebration.
It's also great way to post a couple of pictures of some of the Geospiza team.
 |
| Pictured (l-r): Karen Friery, Sarah Lauer, Stephanie Tatem Murphy, Darrell Reising, Jackie Wright and Jeff Kozlowski. |
 |
| Pictured (l-r): Darrell Reising, Jim Hancock, Sarah Lauer, Stephanie Tatem Murphy, Todd Smith, Jackie Wright and Jeff Kozlowski |
About the activity
After thousands celebrate the Fourth of July and the watch the fireworks, Seattle’s Lake Union can be more trampled fields of chip bags and shining mounds of beer cans than amber waves of grain and purple mountains majesty.
As a member of the biotech community based around Lake Union, Geospiza donned gardening gloves and joined our neighbors for the 5th of July Lake Union Cleanup, a post-fireworks cleaning frenzy hosted by Starbucks, Puget Sound Keeper Alliance and Seattle Public Utilities.
Last year’s inaugural event had more than 700 volunteers collecting 1.15 tons of garbage – one-third of which was recycled. The cleaning event grew out of the community-based efforts to save the fireworks. When the 2010 show was cancelled due to lack of corporate sponsorship, the community launched a fund drive which raised enough money in less than 24hrs.
From April through the end of July, every site across PerkinElmer, including Geospiza, participates in one “For The Better Day” event to fulfill our company mission to improve human and environmental health while providing employees with an opportunity to build stronger teams and communities where we work, live and play.
Friday, June 10, 2011
Sneak Peak: NGS Resequencing Applications: Part I – Detecting DNA Variants
Join us next Wed. June 15 for a webinar on resequencing applications.
Description:
Description:
This webinar will focus on DNA variant detection using Next Generation Sequencing for the applications of targeted and exome resequencing as well as, whole transcriptome sequencing. The presentation will include an overview of each application and its specific data analysis needs and challenges. Topics covered will include Secondary Analysis (alignments, reference choices, variant detection) with a particular emphasis on DNA variant detection as well as multi-sample comparisons. For in depth comparisons of variant detection methods, Geospiza’s cloud-based GeneSifter Analysis Edition software will be used to assess sample data from NCBI’s GEO and SRA. The webinar will also include a short presentation on how these tools can be deployed for both individual researchers as well as through Geospiza’s Partner Program for NGS sequencing service providers.
Details:
Date and time: Wednesday, June 15, 2011 10:00 am
Pacific Daylight Time (San Francisco, GMT-07:00)
Wednesday, June 15, 2011 1:00 pm
Pacific Daylight Time (San Francisco, GMT-07:00)
Wednesday, June 15, 2011 1:00 pm
Eastern Daylight Time (New York, GMT-04:00)
Wednesday, June 15, 2011 6:00 pm
GMT Summer Time (London, GMT+01:00)
Duration: 1 hour
GMT Summer Time (London, GMT+01:00)
Duration: 1 hour
Tuesday, June 7, 2011
DOE's 2011 Sequencing, Finishing, Analysis in the Future Meeting
 |
| Cactus at Bandelier National Monument |
In addition to standard presentations and panel discussions from the genome centers and sequencing vendors (Life Technologies, Illumina, Roche 454, and Pacific Biosciences), and commercial tech talks, this year's meeting included a workshop on hybrid sequence assembly (mixing Illumina and 454 data, or Illumina and PacBio data). I also presented recent work on how 1000 Genomes and Complete Genomics data are changing our thinking about genetics (abstract below).
John McPherson from the Ontario Cancer Research Institute (OICR, a Geospiza client) gave the kickoff keynote. His talk focused on challenges in cancer sequencing. One of those being that DNA sequencing costs are now predominated by instrument maintenance, sample acquisition, preparation, and informatics, which are never included in the $1000 genome conversation. OICR is now producing 17 trillion bases per month and as they, and others, learn about cancer's complexity, the idea of finding single biochemical targets for magic bullet treatments is becoming less likely.
McPherson also discussed how OICR is getting involved clinical cancer sequencing. Because cancer is a genetic disease, measuring somatic mutations and copy number variations will be best for developing prognostic biomarkers. However, measuring such biomarkers in patients in order to calibrate treatments requires a fast turnaround time between tissue biopsy, sequence data collection, and analysis. Hence, McPherson sees IonTorrent and PacBio as the best platforms for future assays. McPherson closed his presentation stating that data integration is the grand challenge. We're on it!
The remaining talks explored several aspects of DNA sequencing ranging from high throughput single cell sample preparation, to sequence alignment and de novo sequence assembly, to education and interesting biology. I especially liked Dan Distal's (New England Biolabs) presentation on the wood eating microbiome of shipworms. I learned that shipworms are actually little clams that use their shells as drills to harvest the wood. Understanding how the bacteria eat wood is important because we may be able to harness this ability for future energy production.
Finally, there was my presentation for which I've included the abstract.
What's a referenceable reference?
The goal behind investing time and money into finishing genomes to high levels of completeness and accuracy is that they will serve as a reference sequences for future research. Reference data are used as a standard to measure sequence variation, genomic structure, and study gene expression in microarray and DNA sequencing assays. The depth and quality of information that can be gained from such analyses is a direct function of the quality of the reference sequence and level of annotation. However, finishing genomes is expensive, arduous work. Moreover, in the light of what we are learning about genome and species complexity, it is worthwhile asking the question whether a single reference sequence is the best standard of comparison in genomics studies.
The human genome reference, for example, is well characterized, annotated, and represents a considerable investment. Despite these efforts, it is well understood that many gaps exist in even the most recent versions (hg19, build 37) [1], and many groups still use the previous version (hg18, build 36). Additionally, data emerging from the 1000 Genomes Project, Complete Genomics, and others have demonstrated that the variation between individual genomes is far greater than previously thought. This extreme variability has implications for genotyping microarrays, deep sequencing analysis, and other methods that rely on a single reference genome. Hence, we have analyzed several commonly used genomics tools that are based on the concept of a standard reference sequence, and have found that their underlying assumptions are incorrect. In light of these results, the time has come to question the utility and universality of single genome reference sequences and evaluate how to best understand and interpret genomics data in ways that take a high level of variability into account.
Todd Smith(1), Jeffrey Rosenfeld(2), Christopher Mason(3). (1) Geospiza Inc. Seattle, WA 98119, USA (2) Sackler Institute for Comparative Genomics, American Museum of Natural History, New York, NY 10024, USA (3) Weill Cornell Medical College, New York, NY 10021, USA
Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, Alkan C, Malig M, Ventura M, Giannuzzi G, Kallicki J, Anderson P, Tsalenko A, Yamada NA, Tsang P, Kaul R, Wilson RK, Bruhn L, & Eichler EE (2010). Characterization of missing human genome sequences and copy-number polymorphic insertions. Nature methods, 7 (5), 365-71 PMID: 20440878
You can obtain abstracts for all of the presentations at the SFAF website.
Friday, May 20, 2011
21st Century Medicine: A Question of Ps
Last Sunday and Monday (5/15, 5/16/11) the Institute for Systems Biology (ISB) held their annual symposium. This year was the 10th annual and focused on "Systems Biology and P4 Medicine."
For those new to P4 medicine, the Ps stand for Personalized, Predictive, Preventative, and Participatory. P4 medicine is about changing our current disease oriented, reactive, approaches to those that prevent disease by increasing the predictive power of diagnostics. Because we are all different, future diagnostics need to be tailored to each individual, which also means individuals need to be more aware of their health and proactively participate in their health care. The vision of P4 medicine is that it will not only dramatically improve the quality of health care, it will significantly decrease health care costs. Hence, some folks add additional Ps to include payment and policy.
P4 medicine is an ambitious goal. In Lee Hood's closing notes he noted four significant challenges that need to be overcome to make P4 medicine a practical reality:
For those new to P4 medicine, the Ps stand for Personalized, Predictive, Preventative, and Participatory. P4 medicine is about changing our current disease oriented, reactive, approaches to those that prevent disease by increasing the predictive power of diagnostics. Because we are all different, future diagnostics need to be tailored to each individual, which also means individuals need to be more aware of their health and proactively participate in their health care. The vision of P4 medicine is that it will not only dramatically improve the quality of health care, it will significantly decrease health care costs. Hence, some folks add additional Ps to include payment and policy.
P4 medicine is an ambitious goal. In Lee Hood's closing notes he noted four significant challenges that need to be overcome to make P4 medicine a practical reality:
- IT challenges. In addition to working on how to transform datasets containing billions of measurements into actionable information, we need to integrate high dimensional data a wide variety of measurement systems. Reduced data will need to be presented in medical records that can be easily accessed and understood by health care providers and participants.
- Education. Students, scientists, doctors, individuals, and policy makers need to learn and develop an understanding of how the networks of interacting proteins and biochemicals that make us healthy or sick are regulated by our genomes and respond to environmental factors.
- Big vs Small Science. Funding agencies are concerned with how to best support the research needed to create the kinds of technologies and approaches that will unlock biology's complexity to develop future diagnostics and efficacious therapies. Large-scale projects conducted over the past 10 years have made it clear that biology is extremely complex. Deciphering this complexity requires that we integrate production-orientated data collection approaches, that develop a data infrastructure, with focused research projects, run by domain experts, that explore specific ideas. The challenge is balancing big and small science to achieve high impact goals.
- Families. Understanding the genetic basis of health and disease requires the research be conducted on samples derived from families rather than randomized populations. Many families are needed to develop critical insights. However, in the U.S. our IRB (Institutional Review Boards) are considered a hinderance to enrolling individuals.
Through the day and half conference numerous presentations explored different aspects of the above challenges. Walter Jessen at Biomarker Commons has created excellent summaries of the first and second day's presentations. For those who like raw data, the #ISB2011P4 hashtag can be used to get the symposium's tweets.
Friday, May 6, 2011
Big News! PerkinElmer Acquires Geospiza
Yesterday, May 5th, PerkinElmer announced that they have acquired Geospiza. This is exciting news and a great opportunity for Geospiza's current customers, future clients, and the company itself.
From numerous tweets, to more formal news coverage, the response has been great. Xconomy, Genome Web, Genetic Engineering & Biotechnology News (GEN), and many others have covered the story and summarized the key points.
What does it mean?
For our customers, we will continue to provide great lab (LIMS) and analysis software with excellent support. Geospiza will continue to operate in Seattle, because Seattle has a vibrant biotech and software technology environment, which has always been a great benefit for the company.
As PerkinElmer is a global organization committed to delivering advanced technology solutions to improve our health and environment, Geospiza will be able to do more interesting things and grow in exciting ways. For that, stay tuned ...
From numerous tweets, to more formal news coverage, the response has been great. Xconomy, Genome Web, Genetic Engineering & Biotechnology News (GEN), and many others have covered the story and summarized the key points.
What does it mean?
For our customers, we will continue to provide great lab (LIMS) and analysis software with excellent support. Geospiza will continue to operate in Seattle, because Seattle has a vibrant biotech and software technology environment, which has always been a great benefit for the company.
As PerkinElmer is a global organization committed to delivering advanced technology solutions to improve our health and environment, Geospiza will be able to do more interesting things and grow in exciting ways. For that, stay tuned ...
Thursday, April 28, 2011
Product Updates: GeneSifter Lab and GeneSifter Analysis Editions
Spring is here and so are new releases of the GeneSifter products. GeneSifter Lab Edition (GSLE) has been bumped up to 3.17 and GeneSifter Analysis Edition (GSAE) is now at 3.7.
What's New?
GSLE - This release includes big features along and a host of improvements. For starters, we added comprehensive inventory tracking. Now, when you configure forms to track your laboratory processes, you can add and track the use of inventory items.
Inventory items are those reagents, kits, tubes, and other bits that are used to prepare samples for analysis. GSLE makes it easy to add these items and their details like barcodes, lot numbers, vendor data, and expiration dates. Items contain arbitrary units so you can track weights as easily as volumes.
When inventory items are used in the laboratory, they can be included in steps. Each time the item is used, the amount to use can be preconfigured and GSLE will do the math for you. When the inventory item's amount drops below a threshold, GSLE can send an email that includes a link for reordering.
In addition to inventory items we increased support for the PacBio RS, and have made Sample Sheet template design completely user configurable. Sample Sheets are those files that contain the samples' names and other information needed for a data collection run. While GSLE always had good sample sheet support, vendor's frequently change formatting and needed data requirements. In some cases a new software release could be required.
The new sample sheet configuration interface eliminates the above problem, and makes it easy for labs to adapt their sheets to changes. A simple web form is used to define formatting rules and the data that will be added. GSLE tags are used to specify data fields and a search interface provides access to all fields in the database. At run time, the sample sheet is filled with the appropriate data.
View the product sheet to see the interfaces and other features.
GSAE - The new release continues to advance GSAE's data analysis capabilities. Specific features include paired-end data analysis for RNA-Seq and DNA re-sequencing applications. We've also improved the ways in which large datasets can be searched, filtered, and queried. Additional improvements include new dashboards to simplify data access and setting up analysis pipelines.
Finally, for those participating in our partner program, we continue to increase the integration between GSLE and GSAE with single sign on and data transfer features.
More details can be found in the product sheet.
What's New?
GSLE - This release includes big features along and a host of improvements. For starters, we added comprehensive inventory tracking. Now, when you configure forms to track your laboratory processes, you can add and track the use of inventory items.
Inventory items are those reagents, kits, tubes, and other bits that are used to prepare samples for analysis. GSLE makes it easy to add these items and their details like barcodes, lot numbers, vendor data, and expiration dates. Items contain arbitrary units so you can track weights as easily as volumes.
When inventory items are used in the laboratory, they can be included in steps. Each time the item is used, the amount to use can be preconfigured and GSLE will do the math for you. When the inventory item's amount drops below a threshold, GSLE can send an email that includes a link for reordering.
In addition to inventory items we increased support for the PacBio RS, and have made Sample Sheet template design completely user configurable. Sample Sheets are those files that contain the samples' names and other information needed for a data collection run. While GSLE always had good sample sheet support, vendor's frequently change formatting and needed data requirements. In some cases a new software release could be required.
The new sample sheet configuration interface eliminates the above problem, and makes it easy for labs to adapt their sheets to changes. A simple web form is used to define formatting rules and the data that will be added. GSLE tags are used to specify data fields and a search interface provides access to all fields in the database. At run time, the sample sheet is filled with the appropriate data.
View the product sheet to see the interfaces and other features.
GSAE - The new release continues to advance GSAE's data analysis capabilities. Specific features include paired-end data analysis for RNA-Seq and DNA re-sequencing applications. We've also improved the ways in which large datasets can be searched, filtered, and queried. Additional improvements include new dashboards to simplify data access and setting up analysis pipelines.
Finally, for those participating in our partner program, we continue to increase the integration between GSLE and GSAE with single sign on and data transfer features.
More details can be found in the product sheet.
Thursday, April 21, 2011
Science, Culture, Policy
 What do science, culture, and policy have in common? In order to improve the quality and affordability of health care, all three have to change. This message is central to Sage Bionetworks’ mission and the theme from this year’s Sage Commons Congress held April 15th and 16th in San Francisco.
What do science, culture, and policy have in common? In order to improve the quality and affordability of health care, all three have to change. This message is central to Sage Bionetworks’ mission and the theme from this year’s Sage Commons Congress held April 15th and 16th in San Francisco.What’s the problem?
Biology is complex. This complexity makes it difficult to understand why some people are healthy and why others get sick. In some cases we have a clear understanding of the biochemical origins of health conditions and their treatments. Unfortunately, most drugs are effective for only a fraction of the people they treat, and in the cases where drugs are effective, their effectiveness is diminished by side effects. The most striking problems being adverse events, which are the sixth leading cause of death in the US.
One way to cost effectively improve health care is to increase the efficacy of treatments in greater numbers of individuals. Also referred to as personalized medicine, the idea is that future treatments are accompanied by diagnostic tests that indicate the treatment's effectiveness. Accomplishing this goal requires that we understand the ways in which drugs affect their specific and non-specific targets with much higher precision. We also need to understand each target’s role in its biological pathway. However, as we attempt to break systems down into pathways and their component parts, a problem emerges. The components participate in multiple pathways and pathways interact with other pathways to form higher-ordered networks, and these interactions vary within individuals.
 |
| LinkedIn or Biology? |
When visualized, biological networks look a lot like LinkedIn, Facebook, or Twitter networks. In these social networks, individuals participate in many groups and have connections to one another. Unlike social networks, which are easy to dissect, our biological networks comprise millions of interactions between proteins, DNA, RNA, chemicals, and microorganisms. Studying these networks requires advanced data collection technologies, computer programs, software systems, and social interactions. Therein lies the rub.
More data isn't enough
Turning the vision of personalized therapies into reality requires a large numbers of scientists who understand the power of global analyses and can work together in research communities. Hence, one part of the Sage mission is to get greater numbers of scientists to adopt new approaches. Another part is getting them to share their data in useful ways.
Accomplishing this goal requires changing the research culture from one that emphasizes individual contributions to one that promotes group participation. Our publish or perish paradigm, combined with publication business models, discourages open-access and data sharing. It also reduces innovation. According to data presented by Aled Edwards, when faced with the opportunity to look at something completely new, we focus on well-known research problems. Why? Because funding is conservative and doing the same thing as your peers has less risk.
Bottom line, we need to take more risk in our research and take more risk sharing data pre and post publication. Taking more risk means we need to trust each other more.
Simply increasing data availability, however, is not enough. We also need to change health care policy. Vicki Seyfert-Margolis (Senior Advisor, Science Innovation and Policy Office of the FDA) gave a presentation (of her views) that helped explain why research expenditures are increasing while the drug pipeline is getting smaller. She described the medical product ecosystem as a large community of individuals from academia, biotech/pharma, device/diagnostic companies, regulators, payors, physicians, and patients, and discussed how all parties need to be involved in changing their practices to improve outcomes. Seyfert-Margolis closed her presentation by focusing on the public’s (patients') role in changing health policy and how, through social media, the public can become more involved and influence direction.
Changing the world is hard
Sage’s mission is ambitious and audacious. Simultaneously tackling three major problems, as Sage is trying to do, has significant risk. So, what can they do to mitigate their risk and improve success? I’ll close by offering a couple of suggestions.
Education is needed - The numbers of researchers who understand the kinds of data that are needed and how to analyze those data and develop network models is small. Further, technology advances keep moving the target, and evaluating models requires additional bench research and others need to be convinced that this is worthwhile. Sage is on the right path by initiating the conversation, but more is needed to increase understanding and evaluating data. Creating a series of blog articles and tutorials would be good first steps. It would help if presentations focused less on final results and more on data collection and analysis processes.
Be Bold - Part of the congress included presentations about a federation experiment in which a group of labs collaborated by openly sharing their data with each other in real time. This is a good step forward, that would be great step forward if the collaboration ran as a publicly open project where anyone could join. Open it up! Success will be clear when a new group, who is unknown to the current community, forms and accomplishes interesting work with their own data and the commons data. A significant step forward, however, is the openness of the meeting itself as all slides and video recordings of the presentations are available. Additionally, twitter and other forms of realtime communication were encouraged.
Accomplishing this goal requires changing the research culture from one that emphasizes individual contributions to one that promotes group participation. Our publish or perish paradigm, combined with publication business models, discourages open-access and data sharing. It also reduces innovation. According to data presented by Aled Edwards, when faced with the opportunity to look at something completely new, we focus on well-known research problems. Why? Because funding is conservative and doing the same thing as your peers has less risk.
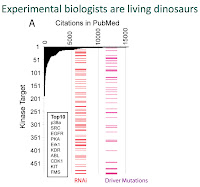 |
| From Edwards' Presentation |
Simply increasing data availability, however, is not enough. We also need to change health care policy. Vicki Seyfert-Margolis (Senior Advisor, Science Innovation and Policy Office of the FDA) gave a presentation (of her views) that helped explain why research expenditures are increasing while the drug pipeline is getting smaller. She described the medical product ecosystem as a large community of individuals from academia, biotech/pharma, device/diagnostic companies, regulators, payors, physicians, and patients, and discussed how all parties need to be involved in changing their practices to improve outcomes. Seyfert-Margolis closed her presentation by focusing on the public’s (patients') role in changing health policy and how, through social media, the public can become more involved and influence direction.
Changing the world is hard
Sage’s mission is ambitious and audacious. Simultaneously tackling three major problems, as Sage is trying to do, has significant risk. So, what can they do to mitigate their risk and improve success? I’ll close by offering a couple of suggestions.
Education is needed - The numbers of researchers who understand the kinds of data that are needed and how to analyze those data and develop network models is small. Further, technology advances keep moving the target, and evaluating models requires additional bench research and others need to be convinced that this is worthwhile. Sage is on the right path by initiating the conversation, but more is needed to increase understanding and evaluating data. Creating a series of blog articles and tutorials would be good first steps. It would help if presentations focused less on final results and more on data collection and analysis processes.
Be Bold - Part of the congress included presentations about a federation experiment in which a group of labs collaborated by openly sharing their data with each other in real time. This is a good step forward, that would be great step forward if the collaboration ran as a publicly open project where anyone could join. Open it up! Success will be clear when a new group, who is unknown to the current community, forms and accomplishes interesting work with their own data and the commons data. A significant step forward, however, is the openness of the meeting itself as all slides and video recordings of the presentations are available. Additionally, twitter and other forms of realtime communication were encouraged.
In closing, I laud Steven Friend and Eric Schadt for founding Sage Bionetworks and pushing the conversation forward. For the past two years the Sage Commons Congress has brought together an amazing and diverse group of participants. The conversation is happening at a critical time because health care needs to change in so many ways. We are at an amazing conversion point with respect to science, technology, and software capabilities that the opportunity for having an impact is high. Geospiza is focused on many aspects of the mission, and we look forward to helping our customers work with their data in new ways. It’s been a pleasure to have the opportunity to participate and help advance progress.
Wednesday, April 6, 2011
Sneak Peak: RNA-Sequencing Applications in Cancer Research: From fastq to differential gene expression, splicing and mutational analysis
Join us next Tuesday, April 12 at 10:00 am PST for a webinar focused on RNA-Seq applications in breast cancer research.
The field of cancer genomics is advancing quickly. News reports from the annual American Association of Cancer Research meeting are indicating that whole genome sequencing studies such as the 50 breast cancer genomes (WashU) are providing more clues about the genes that may be affected in cancer. Meanwhile, the ACLU/Myriad Genetics legal action over genetic testing for breast cancer mutations and disease predisposition continues to move towards the supreme court.
Breast cancer, like many other cancers, is complex. Sequencing genomes is one way to interrogate cancer biology. However, the genome sequence data in isolation does not tell the complete story. The RNA, representing expressed genes, their isoforms, and non-coding RNA molecules, needs to be measured too. In this webinar, Eric Olson, Geospiza's VP of product development and principal designer of GeneSifter Analysis Edition, will explore the RNA world of breast cancer and present how you can explore existing data to develop new insights.
Abstract
Next Generation Sequencing applications allow biomedical researchers to examine the expression of tens of thousands of genes at once, giving researchers the opportunity to examine expression across entire genomes. RNA Sequencing applications such as Tag Profiling, Small RNA and Whole Transcriptome Analysis can identify and characterize both known and novel transcripts, splice junctions and non-coding RNAs. These sequencing based-applications also allow for the examination of nucleotide variant. Next Generation Sequencing and these RNA applications allow researchers to examine the cancer transcriptome at an unprecedented level. This presentation will provide an overview of the gene expression data analysis process for these applications with an emphasis on identification of differentially expressed genes, identification of novel transcripts and characterization of alternative splicing as well as variant analysis and small RNA expression. Using data drawn from the GEO data repository and the Short Read Archive, NGS Tag Profiling, Small RNA and NGS Whole Transcriptome Analysis data will be examined in Breast Cancer.
You can register at the webex site, or view the slides after the presentation.
The field of cancer genomics is advancing quickly. News reports from the annual American Association of Cancer Research meeting are indicating that whole genome sequencing studies such as the 50 breast cancer genomes (WashU) are providing more clues about the genes that may be affected in cancer. Meanwhile, the ACLU/Myriad Genetics legal action over genetic testing for breast cancer mutations and disease predisposition continues to move towards the supreme court.
Breast cancer, like many other cancers, is complex. Sequencing genomes is one way to interrogate cancer biology. However, the genome sequence data in isolation does not tell the complete story. The RNA, representing expressed genes, their isoforms, and non-coding RNA molecules, needs to be measured too. In this webinar, Eric Olson, Geospiza's VP of product development and principal designer of GeneSifter Analysis Edition, will explore the RNA world of breast cancer and present how you can explore existing data to develop new insights.
Abstract
Next Generation Sequencing applications allow biomedical researchers to examine the expression of tens of thousands of genes at once, giving researchers the opportunity to examine expression across entire genomes. RNA Sequencing applications such as Tag Profiling, Small RNA and Whole Transcriptome Analysis can identify and characterize both known and novel transcripts, splice junctions and non-coding RNAs. These sequencing based-applications also allow for the examination of nucleotide variant. Next Generation Sequencing and these RNA applications allow researchers to examine the cancer transcriptome at an unprecedented level. This presentation will provide an overview of the gene expression data analysis process for these applications with an emphasis on identification of differentially expressed genes, identification of novel transcripts and characterization of alternative splicing as well as variant analysis and small RNA expression. Using data drawn from the GEO data repository and the Short Read Archive, NGS Tag Profiling, Small RNA and NGS Whole Transcriptome Analysis data will be examined in Breast Cancer.
You can register at the webex site, or view the slides after the presentation.
Wednesday, March 23, 2011
Translational Bioinformatics
During the week of March 7, I had the pleasure of attending the AMIA’s (American Medical Informatics Association) summit on Translational Bioinformatics (TBI), at the Parc 55 Hotel in San Francisco.
What is Translational Bioinformatics?
Translational Bioinformatics can be simply defined as computer related activities designed to extract clinically actionable information from very large datasets. The field has grown from a need to develop computational methods to work with continually increasing amounts of data and an ever expanding universe of databases.
What is Translational Bioinformatics?
Translational Bioinformatics can be simply defined as computer related activities designed to extract clinically actionable information from very large datasets. The field has grown from a need to develop computational methods to work with continually increasing amounts of data and an ever expanding universe of databases.
As we celebrate the 10th anniversary of completing the draft sequence of the human genome [1,2] we are often reminded that this achievement would transform medicine. The genome would be used to develop a comprehensive catalog of all genes and, through this catalog, we would be able to identify all disease genes and develop new therapies to combat disease. However, since the initial sequence, we have also witnessed an annual decrease in new drugs entering the market place. While progress is being made, it's just not moving at speeds consistent with the excitement produced by the first and “final” drafts [3,4] of the human genome.
What happened?
Biology is complex. Through follow on endeavors, especially with the advent of massively parallel low-cost sequencing, we’ve begun to expose the enormous complexity and diversity of the nearly seven billion genomes that comprise the human species. Additionally, we’ve begun to examine the hundred trillion, or so, microbial genomes that make a human “ecosystem.” A theme that has become starkly evident is our health and wellness is controlled by our genes and how they are modified and expressed in response to environmental factors. Or as described in one slide,
Conferences, like the Joint Summits on Translational Bioinformatics and Clinical Research Informatics, create a forum for individuals working on clinically related computation, bioinformatics and medical informatics problems to come together and share ideas. This year’s meeting was the fourth annual. The TBI meeting had four tracks: 1) Concepts, Tools and Techniques to Enable Integrative Translational Bioinformatics, 2) Integrative Analysis of Molecular and Clinical Measurements, 3) Representing and Relating Phenotypes and Disease for Translational Bioinformatics, and 4) Bridging Basic Science Discoveries and Clinical Practice. To simplify these descriptions, I’d characterize the attendees as participating in five kinds of activities:
What did we learn?
The importance of translational bioinformatics is growing. This year’s summit had about 470 attendees with nearly 300 attending TBI, a 34% growth over 2010 attendance. In addition, to many talks and posters on databases, ontologies, statistical methods, and clinical associations of genes and genotypes, we were entertained by Carl Zimmer’s keynote on the human microbiome.
In his presentation, Zimmer made the case that we need to study human beings as ecosystems. After all our bodies contain between 10 and 100 times more microbial cells than human cells. Zimmer showed how our microbiome becomes populated, from an initially sterile state, through the same niche and succession paradigms that have been demonstrated in other ecosystems. While microbial associations with disease are clear, it is important to note that our microbiome protects us from disease and performs a large number of essential biochemical reactions. In reality our microbiome serves as an additional organ - or two. Thus, to really understand human health, we need to understand the microbiome, so in terms of completing the human ecosystem, we have only peaked at the tip of a very large iceberg, which only gets bigger when we consider bacteriophage and viruses.
Russ Altman closed the TBI portion of the joint summit with his, now annual, year in review. The goals of this presentation were to highlight major trends and advances of the past year, focus on what seems to be important now, and predict what might be accomplished in the coming year. From ~63 papers, 25 were selected for presentation. These were organized into Personal Genomics, Drugs and Genes, Infrastructure for TB, Sequencing and Science, and Warnings and Hope. You can check out the slides to read the full story.
- Creating new statistical models to analyze data
- Integrating diverse data to create new knowledge
- Promoting ontologies and standards
- Developing social media infrastructures and applications
- Using data to perform clinical research and practice
What did we learn?
The importance of translational bioinformatics is growing. This year’s summit had about 470 attendees with nearly 300 attending TBI, a 34% growth over 2010 attendance. In addition, to many talks and posters on databases, ontologies, statistical methods, and clinical associations of genes and genotypes, we were entertained by Carl Zimmer’s keynote on the human microbiome.
In his presentation, Zimmer made the case that we need to study human beings as ecosystems. After all our bodies contain between 10 and 100 times more microbial cells than human cells. Zimmer showed how our microbiome becomes populated, from an initially sterile state, through the same niche and succession paradigms that have been demonstrated in other ecosystems. While microbial associations with disease are clear, it is important to note that our microbiome protects us from disease and performs a large number of essential biochemical reactions. In reality our microbiome serves as an additional organ - or two. Thus, to really understand human health, we need to understand the microbiome, so in terms of completing the human ecosystem, we have only peaked at the tip of a very large iceberg, which only gets bigger when we consider bacteriophage and viruses.
Russ Altman closed the TBI portion of the joint summit with his, now annual, year in review. The goals of this presentation were to highlight major trends and advances of the past year, focus on what seems to be important now, and predict what might be accomplished in the coming year. From ~63 papers, 25 were selected for presentation. These were organized into Personal Genomics, Drugs and Genes, Infrastructure for TB, Sequencing and Science, and Warnings and Hope. You can check out the slides to read the full story.
My take away is that we’ve clearly initiated personal genomics with both clinical and do-it-yourself perspectives. Semantic standards will improve computational capabilities, but we should not hesitate to mine and use data from medical records, participant driven studies, and previously collected datasets in our association studies. Pharmacogenetics/genomics will change drug treatments from one-size-fits-all-benifits-few approaches to specific drugs for stratified populations and multi-drug therapies will become the norm. Deep sequencing continues to reveal deep complexity in our genome, cancer genomes, and the microbiome.
Altman closed with his 2011 predictions. He predicted that consumer sequencing (vs. genotyping) will emerge, cloud computing will contribute to a major biomedical discovery, informatics application to stem cell science will emerge, important discoveries will be made from text mining along with population-based data mining, systems modeling will suggest useful polypharmacy, and immune genomics will emerge as powerful data.
I expect many of these will come true as our [Geospiza's] research and development, and customer engagements are focused on many of the above targets.
References
Consortium efforts (2001). Human genome sequencing issues: Nature, 409 (6288) DOI: Nature, Vol. 409, no 6822 pp. 745-964
1. Nature, Vol. 409, no 6822 pp. 745-964
2. Science, Vol. 291, no. 5507 pp. 1145-1434
3. Nature, Vol. 422, no. 6934 pp. 835-847
4. Science, Vol. 300, no. 5617 pp. 286-290
AMIA - https://www.amia.org
Altman closed with his 2011 predictions. He predicted that consumer sequencing (vs. genotyping) will emerge, cloud computing will contribute to a major biomedical discovery, informatics application to stem cell science will emerge, important discoveries will be made from text mining along with population-based data mining, systems modeling will suggest useful polypharmacy, and immune genomics will emerge as powerful data.
I expect many of these will come true as our [Geospiza's] research and development, and customer engagements are focused on many of the above targets.
References
Consortium efforts (2001). Human genome sequencing issues: Nature, 409 (6288) DOI: Nature, Vol. 409, no 6822 pp. 745-964
1. Nature, Vol. 409, no 6822 pp. 745-964
2. Science, Vol. 291, no. 5507 pp. 1145-1434
3. Nature, Vol. 422, no. 6934 pp. 835-847
4. Science, Vol. 300, no. 5617 pp. 286-290
AMIA - https://www.amia.org
Thursday, March 10, 2011
Sneak Peak: The Next Generation Challenge: Developing Clinical Insights Through Data Integration
Next week (March 14-18, 2011) is CHI's X-Gen Congress & Expo. I'll be there presenting a poster on the next challenge in bioinformatics, also known as the information bottleneck.
You can follow the tweet by tweet action via @finchtalk or #XGenCongress.
In the meantime, enjoy the poster abstract.
The next generation challenge: developing clinical insights through data integration
You can follow the tweet by tweet action via @finchtalk or #XGenCongress.
In the meantime, enjoy the poster abstract.
The next generation challenge: developing clinical insights through data integration
Next generation DNA sequencing (NGS) technologies hold great promise as tools for building a new understanding of health and disease. In the case of understanding cancer, deep sequencing provides more sensitive ways to detect the germline and somatic mutations that cause different types of cancer as well as identify new mutations within small subpopulations of tumor cells that can be prognostic indicators of tumor growth or drug resistance. Intense vendor competition amongst NGS platform and service providers are commoditizing data collection costs making data more assessable. However, the single greatest impediment to developing relevant clinical information from these data is the lack of systems that create easy access to the immense bioinformatics and IT infrastructures needed for researchers to work with the data.
In the case of variant analysis, such systems will need to process very large datasets, and accurately predict common, rare, and de novo levels of variation. Genetic variation must be presented in an annotation-rich, biological context to determine the clinical utility, frequency, and putative biological impact. Software systems used for this work must integrate data from many samples together with resources ranging from core analysis algorithms to application specific datasets to annotations, all woven into computational systems with interactive user interfaces (UIs). Such end-to-end systems currently do not exist, but the parts are emerging.
Geospiza is improving how researchers understand their data in terms of its biological context, function and potential clinical utility, by develop methods that combine assay results from many samples with existing data and information resources from dbSNP, 1000 Genomes, cancer genome databases, GEO, SRA and others. Through this work, and follow on product development, we will produce integrated sensitive assay systems that harness NGS for identifying very low (1:1000) levels of changes between DNA sequences to detect cancerous mutations, emerging drug resistance, and early-stage signaling cascades.
Authors: Todd M. Smith(1), Christoper Mason(2)
(1). Geospiza Inc. Seattle WA 98119, USA.
(2). Weil Cornell Medical College, NY NY 10021, USA
Thursday, March 3, 2011
The Flavors of SNPs
In Blink, Malcolm Gladwell discusses how experts do expert things. Essentially they develop granular languages to describe the characteristics of items, or experiences. Food tasters, for example, use a large and rich vocabulary with scores to describe a food’s aroma, texture, color, taste, and other attributes.
We characterize DNA variation in a similar way
In a previous post, I presented a high level analysis of dbSNP, NCBI’s catalog of human variation. The analysis utilized the VCF (variant call format) file that holds the entire collection of variants and their many annotations. From these data we learned about dbSNP’s growth history, the distribution of variants by chromosome, and additional details such as the numbers of alleles that are recorded for a variant or its structure. We can further classify dbSNP’s variants, like flavors of coffee, by examining the annotation tags that accompany each one.
So, how do they taste?
Each variant in dbSNP can be accompanied by one or more annotations (tags) that define particular attributes, or things we know about a variant. Tags are listed at top of the VCF file in lines that begin with “##INFO.” There 49 such lines. The information about each tag, it’s name, type, and a description is included between angle (<>) brackets. Tag names are coded in alphanumeric values. Most are simple flags, but some include numeric (integer or float) values.
Tags can also be grouped (arbitrarily) into categories to further understand the current state of knowledge about dbSNP. In this analysis I organized 42 of the 49 tags into six categories called: clinical value, link outs, gene structure, bioinformatics issues, population biology (pop. bio.), and 1000 genomes. The seven excluded tags either described house keeping issues (dbSNPBuildID), structural features (RS, VC), non-human readable bitfields (VP), or fields that do not seem to be used or have the same value for every variant (NS [not used], AF [always 0], WGT [always 1]).
 By exploring the remaining 42 tags we assess our current understanding about human variation. For example, approximately 10% of the variants in the database lack tags. For these, we only know that that they have been found in some experiment somewhere. The most common number of tags for a variant is two, and a small number of variants (148,992) have more than ten tags. 88% of variants in the database have between one and ten tags. Put another way, one could say that 40% of the variants are poorly characterized having between zero and two tags, 42% are moderately characterized having between three and six tags, and 16% well characterized, having seven or more tags.
By exploring the remaining 42 tags we assess our current understanding about human variation. For example, approximately 10% of the variants in the database lack tags. For these, we only know that that they have been found in some experiment somewhere. The most common number of tags for a variant is two, and a small number of variants (148,992) have more than ten tags. 88% of variants in the database have between one and ten tags. Put another way, one could say that 40% of the variants are poorly characterized having between zero and two tags, 42% are moderately characterized having between three and six tags, and 16% well characterized, having seven or more tags.
We can add more flavor
 We can also count the numbers of variants that fall into categories of tags. A very small number 135,473 (0.5%) have tags that describe possible clinical value. Clinical value tags are applied to variants that are known diagnostic markers (CLN, CDA), have clinical PubMed citations (PM), exist in the PharmGKB database (TPA), are cited as a mutations from a reputable source (MUT), exist in locus specific databases (LSD), have an attribution from a genome wide association study (MTP), or exist in the OMIM/OMIA database (OM). Interestingly 20,544 variants are considered clinical (CLN), but none are tagged as being “interrogated in a clinical diagnostic assay” (CDA). Also, while I’ve grouped 0.5% of the variants in the clinical value category, the actual number is likely lower, because the tag definitions contain obvious overlaps with other tags in this category.
We can also count the numbers of variants that fall into categories of tags. A very small number 135,473 (0.5%) have tags that describe possible clinical value. Clinical value tags are applied to variants that are known diagnostic markers (CLN, CDA), have clinical PubMed citations (PM), exist in the PharmGKB database (TPA), are cited as a mutations from a reputable source (MUT), exist in locus specific databases (LSD), have an attribution from a genome wide association study (MTP), or exist in the OMIM/OMIA database (OM). Interestingly 20,544 variants are considered clinical (CLN), but none are tagged as being “interrogated in a clinical diagnostic assay” (CDA). Also, while I’ve grouped 0.5% of the variants in the clinical value category, the actual number is likely lower, because the tag definitions contain obvious overlaps with other tags in this category.
2.3 million variants are used as markers on high density genotyping kits (HD) - from the VCF file we don’t know whose kits, just that they're used. The above graph places them in this category, but they are not counted with the clinical value tags, because we only know they are a marker on some kit somewhere. Many variants (37%) contain links to other resources such as 3D structures (S3D), a PubMed Central article (PMC), or somewhere else that is not documented in the VCF file (SLO). Additional details about the kits and links can likely be found in individual dbSNP records.
Why is this important?
When next generation sequencing is discussed, is often accompanied by a conversation about the bioinformatics bottleneck, which deals with the computation phase of data analysis where raw sequences are reduced to quantitative values or lists of variation. Some also discuss a growing information bottleneck that refers to the interpretation of the data. That is, what do those patterns of variation or lists of differentially expressed genes mean? Developing gene models of health and disease and other biological insights requires that assay data can be integrated with other forms of existing information. However, this information is held in a growing number of specialized databases and literature that are scattered and changing rapidly. Integrating such information into systems, beyond simple data links, will require deep characterization of each resource to put assay data into a biological context.
The work described in this and the previous post, provides a short example of the kinds of things that will need to be done with many different databases to increase their utility in aiding in the development of biological models that can be used to link genotype to phenotype.
Further Reading
Sherry ST, Ward M, & Sirotkin K (1999). dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome research, 9 (8), 677-9 PMID: 10447503
1000 Genomes Project Consortium, Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, & McVean GA (2010). A map of human genome variation from population-scale sequencing. Nature, 467 (7319), 1061-73 PMID: 20981092
We characterize DNA variation in a similar way
In a previous post, I presented a high level analysis of dbSNP, NCBI’s catalog of human variation. The analysis utilized the VCF (variant call format) file that holds the entire collection of variants and their many annotations. From these data we learned about dbSNP’s growth history, the distribution of variants by chromosome, and additional details such as the numbers of alleles that are recorded for a variant or its structure. We can further classify dbSNP’s variants, like flavors of coffee, by examining the annotation tags that accompany each one.
So, how do they taste?
Each variant in dbSNP can be accompanied by one or more annotations (tags) that define particular attributes, or things we know about a variant. Tags are listed at top of the VCF file in lines that begin with “##INFO.” There 49 such lines. The information about each tag, it’s name, type, and a description is included between angle (<>) brackets. Tag names are coded in alphanumeric values. Most are simple flags, but some include numeric (integer or float) values.
Tags can also be grouped (arbitrarily) into categories to further understand the current state of knowledge about dbSNP. In this analysis I organized 42 of the 49 tags into six categories called: clinical value, link outs, gene structure, bioinformatics issues, population biology (pop. bio.), and 1000 genomes. The seven excluded tags either described house keeping issues (dbSNPBuildID), structural features (RS, VC), non-human readable bitfields (VP), or fields that do not seem to be used or have the same value for every variant (NS [not used], AF [always 0], WGT [always 1]).
 By exploring the remaining 42 tags we assess our current understanding about human variation. For example, approximately 10% of the variants in the database lack tags. For these, we only know that that they have been found in some experiment somewhere. The most common number of tags for a variant is two, and a small number of variants (148,992) have more than ten tags. 88% of variants in the database have between one and ten tags. Put another way, one could say that 40% of the variants are poorly characterized having between zero and two tags, 42% are moderately characterized having between three and six tags, and 16% well characterized, having seven or more tags.
By exploring the remaining 42 tags we assess our current understanding about human variation. For example, approximately 10% of the variants in the database lack tags. For these, we only know that that they have been found in some experiment somewhere. The most common number of tags for a variant is two, and a small number of variants (148,992) have more than ten tags. 88% of variants in the database have between one and ten tags. Put another way, one could say that 40% of the variants are poorly characterized having between zero and two tags, 42% are moderately characterized having between three and six tags, and 16% well characterized, having seven or more tags.We can add more flavor
 We can also count the numbers of variants that fall into categories of tags. A very small number 135,473 (0.5%) have tags that describe possible clinical value. Clinical value tags are applied to variants that are known diagnostic markers (CLN, CDA), have clinical PubMed citations (PM), exist in the PharmGKB database (TPA), are cited as a mutations from a reputable source (MUT), exist in locus specific databases (LSD), have an attribution from a genome wide association study (MTP), or exist in the OMIM/OMIA database (OM). Interestingly 20,544 variants are considered clinical (CLN), but none are tagged as being “interrogated in a clinical diagnostic assay” (CDA). Also, while I’ve grouped 0.5% of the variants in the clinical value category, the actual number is likely lower, because the tag definitions contain obvious overlaps with other tags in this category.
We can also count the numbers of variants that fall into categories of tags. A very small number 135,473 (0.5%) have tags that describe possible clinical value. Clinical value tags are applied to variants that are known diagnostic markers (CLN, CDA), have clinical PubMed citations (PM), exist in the PharmGKB database (TPA), are cited as a mutations from a reputable source (MUT), exist in locus specific databases (LSD), have an attribution from a genome wide association study (MTP), or exist in the OMIM/OMIA database (OM). Interestingly 20,544 variants are considered clinical (CLN), but none are tagged as being “interrogated in a clinical diagnostic assay” (CDA). Also, while I’ve grouped 0.5% of the variants in the clinical value category, the actual number is likely lower, because the tag definitions contain obvious overlaps with other tags in this category.2.3 million variants are used as markers on high density genotyping kits (HD) - from the VCF file we don’t know whose kits, just that they're used. The above graph places them in this category, but they are not counted with the clinical value tags, because we only know they are a marker on some kit somewhere. Many variants (37%) contain links to other resources such as 3D structures (S3D), a PubMed Central article (PMC), or somewhere else that is not documented in the VCF file (SLO). Additional details about the kits and links can likely be found in individual dbSNP records.
Variants for which something is known about how they map within or near genes account for 42% of the database. Gene structure tags identify variants that result in biochemical consequences such as frameshift (NSF), missense (amino acid change, NSM), nonsense (conversion to sstop codon, NSN), or splice donor and acceptor sequence (DSS, ASS) changes. Coding region variations that do not result in amino changes, or truncated proteins are also identified (SYN, REF). In addition to coding regions, variants can also be found in 5’ and 3’ untranslated regions (U5, U3), proximal and distal to genes (R5, R3), and within introns (INT). Not surprisingly, nearly 37% of the variants in map to introns.
Bioinformatics issues form an interesting category of tags. These include tags that describe a variant’s mapping to different assemblies of the human genome (OTH, CFL, ASP), whether a variant is a unique contig allele (NOC), whether variants have more than two alleles from different submissions (NOV), or if a variant has a genotype conflict (GCF); 22% of the variants are tagged with one or more of these issues. Bioinformatics issues speak to the fact that reference sequence is much more complicated than we like to think.
Because my tag grouping is arbitrary, I’ve also included validated variants (VLD) as a bioinformatics issue. In this case it's a positive issue, because 30% of dbSNP’s have been validated in someway. Of course, a more pessimistic view will see this as 70% are not validated. Indeed, from various communications, dbSNP may have a false positive rate that is on the order of 10%.
The last two categories organize tags by population biology and variants contributed by the 1000 Genomes Project. Population biology tags G5A and G5 describe 5% minor allele frequencies computed by different methods. GNO variants are those that have a genotype available. Over 24 million (85%) variants have been delivered through the 1000 Genome Project's two pilot phases (KGPilot1, KGPilot123). Of these approximately 4 million have been genotyped (PH1). The 1000 genomes category also includes the greatest number of unused tags as this, like everything else we are doing in genomics today, is a work in progress.
Bioinformatics issues form an interesting category of tags. These include tags that describe a variant’s mapping to different assemblies of the human genome (OTH, CFL, ASP), whether a variant is a unique contig allele (NOC), whether variants have more than two alleles from different submissions (NOV), or if a variant has a genotype conflict (GCF); 22% of the variants are tagged with one or more of these issues. Bioinformatics issues speak to the fact that reference sequence is much more complicated than we like to think.
Because my tag grouping is arbitrary, I’ve also included validated variants (VLD) as a bioinformatics issue. In this case it's a positive issue, because 30% of dbSNP’s have been validated in someway. Of course, a more pessimistic view will see this as 70% are not validated. Indeed, from various communications, dbSNP may have a false positive rate that is on the order of 10%.
The last two categories organize tags by population biology and variants contributed by the 1000 Genomes Project. Population biology tags G5A and G5 describe 5% minor allele frequencies computed by different methods. GNO variants are those that have a genotype available. Over 24 million (85%) variants have been delivered through the 1000 Genome Project's two pilot phases (KGPilot1, KGPilot123). Of these approximately 4 million have been genotyped (PH1). The 1000 genomes category also includes the greatest number of unused tags as this, like everything else we are doing in genomics today, is a work in progress.
Why is this important?
When next generation sequencing is discussed, is often accompanied by a conversation about the bioinformatics bottleneck, which deals with the computation phase of data analysis where raw sequences are reduced to quantitative values or lists of variation. Some also discuss a growing information bottleneck that refers to the interpretation of the data. That is, what do those patterns of variation or lists of differentially expressed genes mean? Developing gene models of health and disease and other biological insights requires that assay data can be integrated with other forms of existing information. However, this information is held in a growing number of specialized databases and literature that are scattered and changing rapidly. Integrating such information into systems, beyond simple data links, will require deep characterization of each resource to put assay data into a biological context.
The work described in this and the previous post, provides a short example of the kinds of things that will need to be done with many different databases to increase their utility in aiding in the development of biological models that can be used to link genotype to phenotype.
Further Reading
Sherry ST, Ward M, & Sirotkin K (1999). dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome research, 9 (8), 677-9 PMID: 10447503
1000 Genomes Project Consortium, Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, & McVean GA (2010). A map of human genome variation from population-scale sequencing. Nature, 467 (7319), 1061-73 PMID: 20981092
Tuesday, March 1, 2011
Data Analysis for Next Generation Sequencing: Challenges and Solutions
Join us next Tuesday, March 8 at 10:00 am Pacific Time for a webinar on Next Gen sequence data analysis.
Register Today!
http://geospizaevents.webex.com
Please note the previous post had Wed. listed for the day. The correct day is Tue.
Wednesday, February 16, 2011
Sneak Peak: ABRF and Software Systems for Clinical Research
The Association for Biomedical Research Facilities conference begins this weekend (2/19) with workshops on Saturday and sessions Sunday through Tuesday. This year's theme is: Technologies to Enable Personalized Medicine, and appropriately a team from Geospiza will be there at our booth and participating in scientific sessions.
I will be presenting a poster entitled, "Clinical Systems for Cancer Research" (abstract below). In addition to great science and technology ABRF has a large number of tweeting participants including @finchtalk. You can follow along using the #ABRF and (or) #ABRF2011.
Abstract
Geospiza is transforming the above scenario from vision into reality in several ways. The Company’s GeneSifter platform utilizes scalable data management technologies based on open-source HDF5 and BioHDF technologies to capture, integrate, and mine raw data and analysis results from DNA, RNA, and other high-throughput assays. Analysis results are integrated and linked to multiple repositories of information that include variation, expression, pathway, and ontology databases to enable discovery process and support verification assays. Using this platform and RNA-Sequencing and Genomic DNA sequencing from matched tumor/normal samples, we were able to characterize differential gene expression, differential splicing, allele specific expression, RNA editing, somatic mutations and genomic rearrangements as well as validate these observations in a set of patients with oral and other cancers.
By: Todd Smith (1), N. Eric Olson (1), Rebecca Laborde (3), Christopher E Mason (2), David Smith (3): (1) Geospiza, Inc., Seattle WA. (2) Weil Cornell Medical College, NY NY.(3) Mayo Clinic, Rochester MN.
Friday, February 11, 2011
Variant Analysis and Sequencing Labs
Yesterday and two weeks ago, Geospiza released two important news items. The first announced PerkinElmer's section of the GeneSifter® Lab and Analysis systems to support their new DNA sequencing service. The second was an announcement of our new SBIR award to improve variant detection software.
Why are these important?
The PerkinElmer news is another validation of the fact that software systems need to integrate laboratory operations and scientific data analysis in ways that go deeper than sending computing jobs to a server (links below). To remain competitive, service labs can no longer satisfy customer needs by simply delivering data. They must deliver a unit of information that is consistent with the experiment, or assay, that is being conducted by their clients. PerkinElmer recognizes this fact along with our many other customers who participate in our partner program and work with our LIMS (GSLE) and Analysis (GSAE) systems to support their clients doing Sanger sequencing, Next Gen sequencing, or microarray analysis.
The SBIR news communicates our path toward making the units of information that go with whole genome sequencing, targeted sequencing, exome sequencing, allele analysis, and transcriptome sequencing as rich as possible. Through the project, we will add new applications to our analysis capabilities that solve the three fundamental problems related to data quality analysis, information integration, and the user experience.
Further Reading
Geospiza wins $1.2 M grant to add new DNA variant application to GeneSifter software.
Why are these important?
The PerkinElmer news is another validation of the fact that software systems need to integrate laboratory operations and scientific data analysis in ways that go deeper than sending computing jobs to a server (links below). To remain competitive, service labs can no longer satisfy customer needs by simply delivering data. They must deliver a unit of information that is consistent with the experiment, or assay, that is being conducted by their clients. PerkinElmer recognizes this fact along with our many other customers who participate in our partner program and work with our LIMS (GSLE) and Analysis (GSAE) systems to support their clients doing Sanger sequencing, Next Gen sequencing, or microarray analysis.
The SBIR news communicates our path toward making the units of information that go with whole genome sequencing, targeted sequencing, exome sequencing, allele analysis, and transcriptome sequencing as rich as possible. Through the project, we will add new applications to our analysis capabilities that solve the three fundamental problems related to data quality analysis, information integration, and the user experience.
- Through advanced data quality analysis we can address the question, if we see a variant, can we understand wether the difference is random noise, a systematic error, or biological signal?
- Information integration will help scientists and clinical researchers quickly add biological context to their data.
- New visualization and annotation interfaces will help individuals explore the high-dimensional datasets resulting from the 100's and 1000's of samples needed to develop scientific and clinical insights.
Further Reading
Geospiza wins $1.2 M grant to add new DNA variant application to GeneSifter software.
Tuesday, February 8, 2011
AGBT 2011
More.
That's how I describe this year's conference.
Feel free to add more comments.
That's how I describe this year's conference.
- More attendees
- More data
- More genomes
- More instruments
- More tweeters
- More tweeting controversy
- More software
- More ...
Tuesday, February 1, 2011
Sneak Peak: AGBT and the Next Gen Software Challenge
The Advances in Genome Biology and Technology meeting begins Thursday (2/3) morning with pre-conference workshops on Wed. I will be there representing Geospiza and presenting a poster entitled, "The Next Gen Software Challenge: Integrating Diverse Assay-Seq and Validation Systems" (abstract below)
In addition to great science and technology AGBT has a large number of tweeting participants including @finchtalk. You can follow along using the #AGBT and (or) #AGBT2011.
Abstract
By the end of 2011 we will know the DNA sequences for 30,000 human genomes. Understanding how the variation between these genomes affects phenotype at a molecular level, requires that future research projects integrate genome sequences with data from multiple ultra-high throughput assays obtained from large sample populations. Further insights are gained when these data are combined with additional information from external databases and data resources.Geospiza is meeting the above challenges with its GeneSifter® platform and underlying open-source HDF5 and BioHDF technologies. Our approach centralizes and structures data in a scalable way so that features can be queried across many samples and between different assays to quickly add context to genome sequences as they are collected. Unlike approaches that rely on collections of flat files, specialized software tool kits, isolated genome-browsers, and independent statistical tool environments, GeneSifter integrates common components into a system that provides researchers with a rich, easy to use, environment to explore their data and develop clinical insights.
Friday, January 21, 2011
dbSNP, or is it?
dbSNP is NCBI’s catalog of DNA variation. While the SNP in the name implies a focus on Single Nucleotide Polymorphisms, dbSNP is far more comprehensive and includes length variants, mutations, and a plethora of annotations that characterize over 75 million variants from 89 organisms.
 The entire collection of human variants - minus 1,617,401 - can be obtained in a single VCF file. VCF stands for Variant Call Format ; it is a standard created by the 1000 Genomes project [2] to list and annotate genotypes. The vcard format also uses the “vcf” file extension . Thus, genomic vcf files have cute business card icons on Macs and Windows. Bioinformatics is fun that way.
The entire collection of human variants - minus 1,617,401 - can be obtained in a single VCF file. VCF stands for Variant Call Format ; it is a standard created by the 1000 Genomes project [2] to list and annotate genotypes. The vcard format also uses the “vcf” file extension . Thus, genomic vcf files have cute business card icons on Macs and Windows. Bioinformatics is fun that way.

 So, what can we learn? We can use the dbSNPBuildNumber annotation to observe dbSNP’s growth. Cumulative growth shows that the recent builds have contributed the majority of variants. In particular, the 1000 Genomes project’s contributions account for close to 85% of the variants in dbSNP[2]. Examining the data by build number also shows that a build can consist of relatively few variants. There are even 369 variants from a future 133 build.
So, what can we learn? We can use the dbSNPBuildNumber annotation to observe dbSNP’s growth. Cumulative growth shows that the recent builds have contributed the majority of variants. In particular, the 1000 Genomes project’s contributions account for close to 85% of the variants in dbSNP[2]. Examining the data by build number also shows that a build can consist of relatively few variants. There are even 369 variants from a future 133 build.
 In dbSNP, variants are described in four ways: SNP, INDEL, MIXED, and MULTI-BASE. The vast majority (82%) are SNPs, with INDELs (INsertions and DELetions) forming a large second class. When MIXED and MULTI-BASE variants are examined, they do not appear to look any different than INDELS, so I am not clear on the purpose of this tag. Perhaps it is there for bioinformatics enjoyment, because you have to find these terms in the 30MM lines; they are not listed in the VCF header. It was also interesting to learn that the longest indels are on the order of a 1000 bases.
In dbSNP, variants are described in four ways: SNP, INDEL, MIXED, and MULTI-BASE. The vast majority (82%) are SNPs, with INDELs (INsertions and DELetions) forming a large second class. When MIXED and MULTI-BASE variants are examined, they do not appear to look any different than INDELS, so I am not clear on the purpose of this tag. Perhaps it is there for bioinformatics enjoyment, because you have to find these terms in the 30MM lines; they are not listed in the VCF header. It was also interesting to learn that the longest indels are on the order of a 1000 bases.
 The VCF file contains both reference and variant sequences at each position. The variant sequence can list multiple bases, or sequences, as each position can have multiple alleles. While nearly all variants are a single allele, about 3% have between two and 11 alleles. I assume the variants with the highest numbers of alleles represent complex indel patterns.
The VCF file contains both reference and variant sequences at each position. The variant sequence can list multiple bases, or sequences, as each position can have multiple alleles. While nearly all variants are a single allele, about 3% have between two and 11 alleles. I assume the variants with the highest numbers of alleles represent complex indel patterns.
 Given that the haploid human genome contains approximately 3.1 billion bases, the variants in dbSNP represent nearly 1% of the available positions. Their distribution across the 22 autosomes, as defined by the numbers of positions divided by the chromosome length, is fairly constant around 1%. The X and Y chromosomes and their PARs are, however much lower, with ratios of 0.45%, 0.12%, and 0.08% respectively. A likely reason for this observation is that sampling of sex chromosomes in current sequencing projects is low relative to autosomes. The number of mitochondria variants, as a fraction of genome length, is, on the other hand much higher at 3.95%.
Given that the haploid human genome contains approximately 3.1 billion bases, the variants in dbSNP represent nearly 1% of the available positions. Their distribution across the 22 autosomes, as defined by the numbers of positions divided by the chromosome length, is fairly constant around 1%. The X and Y chromosomes and their PARs are, however much lower, with ratios of 0.45%, 0.12%, and 0.08% respectively. A likely reason for this observation is that sampling of sex chromosomes in current sequencing projects is low relative to autosomes. The number of mitochondria variants, as a fraction of genome length, is, on the other hand much higher at 3.95%.
It is important to point out that the 1% level of variation is simply a count of all currently known positions where variation can occur in individuals. These data are compiled from many individuals. On a per individual basis, the variation frequency is much lower with each individual having between three and four million differences when their genome is compared to the other people's genomes.
Previously, I discussed how the numbers of biological information repositories are growing each year. Nucleic Acids Research now tracks over 1300 databases that contain specialized subsets of DNA, RNA, protein sequences, and other kinds of biochemical data along with annotations (metadata) that can be mined or searched to aid our scientific research.
Most of these resources have descriptions and publications describing high-level details about their mission with respect to what kinds of data are stored and how the resource can be used. While useful, these descriptions are typically out of date, because, like everything else in information science, each repository is undergoing significant growth in terms of data stored and the data’s annotations. As we contemplate how to use these resources in integrated analyses we need methods to summarize their content and extract data in global ways.
As an example, let’s consider dbSNP. According to dbSNP’s build history the first release was in Dec 1, 1998. Build 2, 9 days later, had 11 new SNPs added from the debnick’s 981209.dat file. Now, dbSNP is 12 years old and is at build 132. The term build is a software way of saying version.
Several methods can be used to access dbSNP data. These include multiple web interfaces at NCBI and flat files that hold entire datasets in XML, SQL tables, and VCF formats. When first published, in 1999 [1], dbSNP contained 4713 variants and few annotations. As of build 132, the human specific database contains either 30,443,455 (release notes) or 28,826,054 variants (VCF file). Why the discrepancy? Share your idea with a comment.
What else can we learn about human DNA variation from dbSNP’s VCF file?
 The entire collection of human variants - minus 1,617,401 - can be obtained in a single VCF file. VCF stands for Variant Call Format ; it is a standard created by the 1000 Genomes project [2] to list and annotate genotypes. The vcard format also uses the “vcf” file extension . Thus, genomic vcf files have cute business card icons on Macs and Windows. Bioinformatics is fun that way.
The entire collection of human variants - minus 1,617,401 - can be obtained in a single VCF file. VCF stands for Variant Call Format ; it is a standard created by the 1000 Genomes project [2] to list and annotate genotypes. The vcard format also uses the “vcf” file extension . Thus, genomic vcf files have cute business card icons on Macs and Windows. Bioinformatics is fun that way. The dbSNP VCF file is a convenient way to get a global view of the database. Variants are listed by chromosome and position. Each position also lists the NCBI rs ID, reference, and variant sequence(s). Chromosomes include the 22 autosomes, X, Y, MT (mitochondria) and PAR (Pseudoautosomal Regions[3]). The last column (INFO) is the most interesting. It can contain up to 49 specific annotations that describe a variant’s type, its origin, biological features, population characteristics, and whether is it linked to other resources. Many of the annotations are tags, but some contain additional information. Between the different tags and information within tags, there are over 50 ways to describe a variant. One of the most obfuscated annotations is a 12-byte bitfield, which must be decoded to understand. No worry, much of its information appears to be repeated in the readable annotations. Others have also noted that the bitfield is overly clever.
Digging deeper
We can use the VCF file to learn a great deal about dbSNP and the biology of human variation. But, you cannot do this by reading the file. It is over 30 million lines long! You’re also not going to be able to analyze these data in ExcelTM, so you need to put the data in some kind of database or binary file format. I used HDF5 and pytables to put the data into essentially a 30MM row by 50+ column table that can be efficiently queried using simple python scripts.

 So, what can we learn? We can use the dbSNPBuildNumber annotation to observe dbSNP’s growth. Cumulative growth shows that the recent builds have contributed the majority of variants. In particular, the 1000 Genomes project’s contributions account for close to 85% of the variants in dbSNP[2]. Examining the data by build number also shows that a build can consist of relatively few variants. There are even 369 variants from a future 133 build.
So, what can we learn? We can use the dbSNPBuildNumber annotation to observe dbSNP’s growth. Cumulative growth shows that the recent builds have contributed the majority of variants. In particular, the 1000 Genomes project’s contributions account for close to 85% of the variants in dbSNP[2]. Examining the data by build number also shows that a build can consist of relatively few variants. There are even 369 variants from a future 133 build.  In dbSNP, variants are described in four ways: SNP, INDEL, MIXED, and MULTI-BASE. The vast majority (82%) are SNPs, with INDELs (INsertions and DELetions) forming a large second class. When MIXED and MULTI-BASE variants are examined, they do not appear to look any different than INDELS, so I am not clear on the purpose of this tag. Perhaps it is there for bioinformatics enjoyment, because you have to find these terms in the 30MM lines; they are not listed in the VCF header. It was also interesting to learn that the longest indels are on the order of a 1000 bases.
In dbSNP, variants are described in four ways: SNP, INDEL, MIXED, and MULTI-BASE. The vast majority (82%) are SNPs, with INDELs (INsertions and DELetions) forming a large second class. When MIXED and MULTI-BASE variants are examined, they do not appear to look any different than INDELS, so I am not clear on the purpose of this tag. Perhaps it is there for bioinformatics enjoyment, because you have to find these terms in the 30MM lines; they are not listed in the VCF header. It was also interesting to learn that the longest indels are on the order of a 1000 bases.  The VCF file contains both reference and variant sequences at each position. The variant sequence can list multiple bases, or sequences, as each position can have multiple alleles. While nearly all variants are a single allele, about 3% have between two and 11 alleles. I assume the variants with the highest numbers of alleles represent complex indel patterns.
The VCF file contains both reference and variant sequences at each position. The variant sequence can list multiple bases, or sequences, as each position can have multiple alleles. While nearly all variants are a single allele, about 3% have between two and 11 alleles. I assume the variants with the highest numbers of alleles represent complex indel patterns. Given that the haploid human genome contains approximately 3.1 billion bases, the variants in dbSNP represent nearly 1% of the available positions. Their distribution across the 22 autosomes, as defined by the numbers of positions divided by the chromosome length, is fairly constant around 1%. The X and Y chromosomes and their PARs are, however much lower, with ratios of 0.45%, 0.12%, and 0.08% respectively. A likely reason for this observation is that sampling of sex chromosomes in current sequencing projects is low relative to autosomes. The number of mitochondria variants, as a fraction of genome length, is, on the other hand much higher at 3.95%.
Given that the haploid human genome contains approximately 3.1 billion bases, the variants in dbSNP represent nearly 1% of the available positions. Their distribution across the 22 autosomes, as defined by the numbers of positions divided by the chromosome length, is fairly constant around 1%. The X and Y chromosomes and their PARs are, however much lower, with ratios of 0.45%, 0.12%, and 0.08% respectively. A likely reason for this observation is that sampling of sex chromosomes in current sequencing projects is low relative to autosomes. The number of mitochondria variants, as a fraction of genome length, is, on the other hand much higher at 3.95%.It is important to point out that the 1% level of variation is simply a count of all currently known positions where variation can occur in individuals. These data are compiled from many individuals. On a per individual basis, the variation frequency is much lower with each individual having between three and four million differences when their genome is compared to the other people's genomes.
Clearly, from the original publications and current dbSNP reports, the database has grown significantly and is much richer and undergoing changes in ways that cannot be easily communicated through traditional publication methods. The challenge going forward is developing software that can adapt to these changes to evaluate resources and use their content in effective ways.
Next time I’ll discuss the annotations.
References and notes:
1. Sherry ST, Ward M, & Sirotkin K (1999). dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome research, 9 (8), 677-9 PMID: 10447503
1. Sherry ST, Ward M, & Sirotkin K (1999). dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome research, 9 (8), 677-9 PMID: 10447503
2. 1000 Genomes Project (1KG) is an endeavor to characterize DNA sequence variation in humans. Visit the website at www.1000genomes.org or read the paper: 1000 Genomes Consortium. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073.
3. PAR - PseudoAutosomal Regions - are located at the ends of the X and Y chromosome. The DNA in these regions can recombine between the sex chromosomes, hence PAR genes demonstrate autosomal inheritance patterns, rather than sex inheritance patterns. You can read more at http://en.wikipedia.org/wiki/Pseudoautosomal_region
3. PAR - PseudoAutosomal Regions - are located at the ends of the X and Y chromosome. The DNA in these regions can recombine between the sex chromosomes, hence PAR genes demonstrate autosomal inheritance patterns, rather than sex inheritance patterns. You can read more at http://en.wikipedia.org/wiki/Pseudoautosomal_region
Subscribe to:
Posts (Atom)